| Strand NGS v3.3 Release Announcement |
| Strand Life Sciences is happy to announce the release of Strand NGS v3.3.
This release focuses on two aspects - experiment level Quality Control checks to identify any substandard samples and enabling diverse analysis in Single Cell Transcriptome studies.
|
|
|
| Experiment level Quality Control checks |
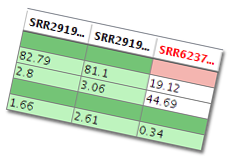
| Identify substandard samples, if any - by introducing a QC Check Manager, we enable the flagging of substandard samples in any alignment experiment based on the standards that you can configure. This helps you get a comprehensive picture on the quality of all the samples in a given alignment run before using them for further analysis. |
|
|
| Enabling Single Cell Transcriptome studies |
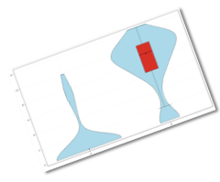
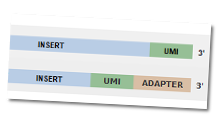
| We have now added a number of features like cellular demultiplexing, tagging cells that have sub-optimal number of reads, violin plots for depicting gene expression distribution, data distribution overlay in heatmap legend, which aid in exploring your Single Cell data sets better than ever.
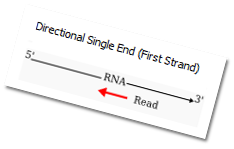
Support for Directional Single End First Strand libraries makes our library layout support as comprehensive as it can get. Also, our algorithm for detection of novel splices during alignment is now highly enhanced. |